Machine learning projects require a different approach compared to traditional software development. After working on numerous ML projects, I’ve compiled practical tips to streamline the research and development process. This guide doesn’t require specific ML knowledge but focuses on effective workflows and organizational strategies.
I tried to find online mainly through Google to find articles for similar purposes but could not anything useful before completing the initial draft. I wrote this by myself based on my own experience and domain of knowledge. Later on, I found more articles with similar purposes while reading Lillian Weng’s blog1. I amended this post after the initial version by trying to map add additional info from other blog posts onto my current article’s structure. If my tips contain refernce to another blog, this means it really important since we all think something is useful although I’m nobody and authors from other blog posts are really good researchers. If I don’t have certain tips, I need to learn. Last but not least, if I have some original ideas, please verify before you adopt.
Later on after I initially published this blog post, I started to find out some really good papers that are really professional, meaning their methods are way better than mine in terms of detail. I’ll also try to add those into this blog post so that readers could benefit from specific tools within each tip. Some of the good papers including BenchMARL2. I recommend you to check it out.
Development: Setting Up for Success
The development phase focuses on creating functional code and establishing proper architecture. During this stage, we’re primarily concerned with generating code that works and can serve as a foundation for our research. This means defining architectures thoughtfully and managing our codebase efficiently from the start.
Decouple Code, Data, and Configuration
Unlike traditional software development, ML projects involve three distinct elements: your code (algorithms and implementation), your data (training and evaluation datasets), and your configuration (hyperparameters and experiment settings).
Keeping these separate makes your project more manageable. Particularly, configurations should be stored in separate files (YAML, JSON, etc.) rather than hardcoded. This separation facilitates easier hyperparameter tuning, cleaner experiment tracking, more reproducible results, and simplified collaboration.
For example, instead of hardcoding parameters like learning_rate = 0.001, use configuration files that separate these decisions from your implementation code. This decoupling proves especially valuable during hyperparameter tuning phases, as you can modify configurations without touching your core algorithm implementations.
Here’s a concrete example of how this separation might look in practice:
project/
├── code/
│ ├── models/
│ ├── data_loaders/
│ ├── trainers/
│ └── evaluation/
├── configs/
│ ├── model_configs/
│ │ ├── resnet50.yaml
│ │ └── transformer.yaml
│ ├── training_configs/
│ │ ├── adam_lr0001.yaml
│ │ └── sgd_cosine.yaml
│ └── experiment_configs/
│ ├── exp_001.yaml
│ └── exp_002.yaml
├── data/
│ ├── raw/
│ ├── processed/
│ └── splits/
└── results/
├── checkpoints/
├── logs/
└── visualizations/This structure allows you to mix and match configurations easily. For instance, you might combine resnet50.yaml with adam_lr0001.yaml for one experiment, then quickly switch to using transformer.yaml with the same optimizer config for comparison.
Daniel Seshi’s article3 presents argparse for inline arguments parsing, reduced inline arguments parsing by setting default values, customized shell script, using json or yaml.
Version Control is Non-Negotiable
Git should be central to your development process. Beyond standard version control benefits, ML projects particularly benefit from tracking experiment configurations, documenting model architecture changes, enabling collaboration on complex models, and creating branches for different approaches. Daniel Seshi’s article3 put git as the most fundamental tip for research management.
For ML projects, commit configuration changes separately from code changes, use meaningful commit messages that reference experiment IDs, consider git-lfs for larger files (though not for full datasets), and document environment dependencies alongside code.
A particularly effective git workflow for ML research involves:
# When starting a new experiment approach
git checkout -b feature/attention-mechanism
# After implementing and testing
git add src/models/attention.py
git commit -m "Add self-attention mechanism to encoder"
# When updating configs for a specific experiment
git add configs/exp_attn_001.yaml
git commit -m "Add config for experiment EXP-A001 testing attention with lr=0.0005"
# When experiment yields useful results
git checkout main
git merge feature/attention-mechanismPro tip: Create a .gitignore file that explicitly excludes large datasets, model checkpoints, and generated visualizations while keeping their directory structure. Include patterns like:
# Ignore datasets
data/raw/*
data/processed/*
!data/raw/.gitkeep
!data/processed/.gitkeep
# Ignore checkpoints but keep structure
results/checkpoints/*
!results/checkpoints/.gitkeep
# Allow small sample data for tests
!data/raw/samples/This approach prevents accidentally committing large files while maintaining directory structure and preserving small test samples for CI/CD processes.
Daniel Seshi’s article3 argues to use branches for evaluation code, which I believe is important. What I found is that it would be difficult to get research code in sync with git branches. For example, if I have some branches that are based on the current main head for experimental purpose. When experimenting with another batch of experiments, I need to modify main head first before creating alternative branches or maybe I need to fork a branch to tweak the main head, then based on the modified branch from current main head, I could create more branches for experiment. The reason I mention this is because this git based approach requires you to learn deeper for git, so that you could find a sweet spot for research management.
My argument is that for code and logic related modifications, it worths creating new branches. As to me it is more straightforward that certain code files are changed for a specific purpose that could be considered as a fork. For hyperparameter tuning purpose that happens within the config file, my idea is to put that value in the companion log file such that those optimized values could be cherrypicked from the log file, as a large part of experiments are discarded for the suboptimal results, thus it is not worth committing those hyperparameter value changes.
Env Setup🆕
Daniel Seshi’s article3 relies on pythonenv, requirements.txt, sync those plaintext info with git. In addition, the code repo could also be reused as modules by setting a setup.py, so that other projects could refer to this code. Later on, he switched to conda for the modifications of environments, so my suggestions would be the same. One thing is that one needs to keep an environment setup script to make sure that it could be reused further.
Data Collection and Experiment Management
One most important thing to mention is to backup those experimental data that are not pushed to git server. You could run some commands or manually copy those info to another hard drive. I suggest setting up once and just let the task scheduler to do certain tasks.
Comprehensive Logging
Logging goes beyond git commits. You need to capture training metrics at regular intervals, model states and checkpoints, environment information, data preprocessing details, random seeds, and hardware utilization information.
Create log directories that include the exact code version used (snapshot of key files), configuration files, performance metrics, model checkpoints at intervals, and validation results. This comprehensive logging enables you to reconstruct any experiment later, which proves invaluable when you need to revisit work months after completion.
Random Tags for Experiment Identification
As your experiments multiply, descriptive naming becomes unwieldy. Long descriptive names like lr0.001_batch32_dropout0.5_augTrue_... quickly become unmanageable. I’ve found this becomes particularly problematic when running dozens or hundreds of experiments with slight variations in parameters.
Instead, generate random tags for each experiment such as exp_b4f29a, then map these tags to full configurations in a separate index. This approach creates cleaner filenames, makes experiment references shorter, avoids filename length limitations, and reduces the chance of naming conflicts. When discussing results with colleagues, these short identifiers also simplify communication.
Daniel Seshi’s article3 suggests info including date and time when the experiment executes, arguments parameter info in addition to the random tags. It should be noted that those naming rules are applied to newly created folders.
Saving Models with pickle
Pickle is handy to save trained machine learning models for testing purpose and for data backup goal. I suggest you to save a dict that contains all info including neural network parameters, certain arguments that could help you to reload those models back during the inference phrase. It should be noted that sometimes Pickle may not work in a cross-platform manner, thus I recommend you to do some experiments to see if the pickle file works or not, otherwise this is going to cause a major problem as you cannot load your trained neural networks.
Experiment Scheduling
For running multiple experiments, a scheduler is invaluable. While tools like Slurm exist, a simple custom script can provide more flexibility for personal projects. I’ve found that custom scheduling solutions often better fit the specific needs of ML research workflows, especially when working on personal machines or small clusters.
A basic approach using lock files works well for many scenarios. Create a directory for experiment locks, and when launching an experiment, check the number of lock files. If below your limit, create a new lock file and start the experiment. When an experiment completes, remove its lock file, and new experiments wait until slots are available. My implementation uses a simple script that manages lock files properly—creating them when experiments start, monitoring them with watchdog, and deleting them when tasks complete.
This simple system allows you to queue experiments to run sequentially, maximize resource utilization, avoid manual monitoring, and maintain system stability without complex infrastructure. While this approach doesn’t aim for sophisticated multi-processing capabilities, it efficiently manages sequential tasks with different parameter configurations, which covers most research needs. For truly parallel processing needs, you might want to explore more robust solutions, but I’ve found this lock-file approach strikes an excellent balance between simplicity and functionality.
Visualization Strategies
Terminal Monitoring
The terminal remains a powerful tool for tracking progress. Use tqdm for progress bars that show real-time completion status. I’ve found tqdm particularly useful when running numerous experiments since it provides clear visual feedback on each task’s progress. For enhanced monitoring, try splitting terminals horizontally (iTerm2) to observe multiple experiments simultaneously. This setup creates a dashboard-like view with many rows displaying different tasks. Color-coding output for different experiment types helps distinguish between runs, and displaying key metrics in real-time gives you immediate feedback on performance.
The following code could be used to as a demo on what it looks like for monitoring multiple sessions within a terminal window:
#! /usr/bin/env python3
import tqdm, time
for ep in range(100):
pbar = tqdm.tqdm(range(100), desc=f"Epoch {ep:03d}")
for i in pbar:
# Simulate some metrics
loss = 1.0 - (i / 100)
accuracy = i / 100
pbar.set_postfix(loss=f"{loss:.2f}", acc=f"{accuracy:.2%}")
time.sleep(0.1)By running the code in multiple horizontal splitted window, the following screenshot presents what exactly to monitor for each task:
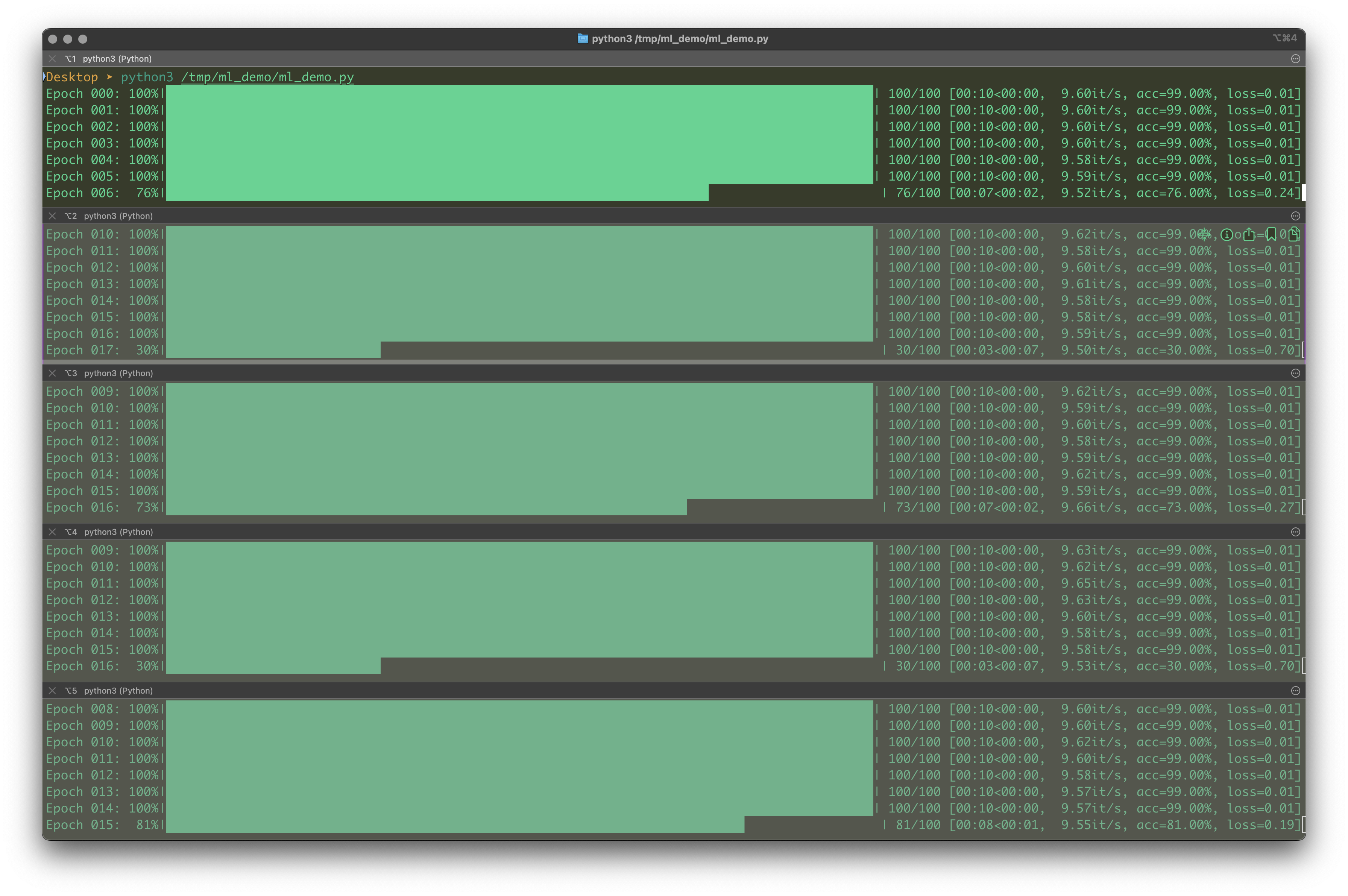
The image above from a Terminal app specifically iTerm2 shows just an example on how to monitor the progresses of multiple Python processes on a Mac. I recall that if you use Tmux on any platform, this kind of setup could be easily replicated.
It should be noted that in my workflow, I would do additional things. First, I’d like to print out key parameters so that I know the process is configurated correctly, I have time to abort current process if something is wrong. Secondly, I would put lots of emojis for example Epoch or Episode could be replaced by #️⃣, you could add 🔄 to indicate whether certain target networks are updated or not. The idea is to add more info onto the terminal so that you know what is going on.
Tensorboard for Comprehensive Tracking
Tensorboard excels at visualizing learning curves, gradient distributions, model architectures, hyperparameter comparisons, and embedding spaces. The interactive interface lets you zoom, filter, and compare runs dynamically. Setting up Tensorboard with appropriate tags makes comparing experiments much easier, especially when dealing with dozens of related runs with subtle parameter differences.
Matplotlib for Custom Visualizations
For specific analysis, create custom visualizations with matplotlib. In machine learning research, visualization is essential for understanding what’s happening with your models. You can overlay predictions on input data, compare multiple model outputs side by side, visualize attention maps or feature importance, and annotate figures with key parameters. These custom visualizations often reveal insights that automated tools might miss. For prototyping especially, I rely heavily on matplotlib to quickly generate informative visualizations that help guide next steps.
Include relevant configuration details directly in figures through descriptive titles and annotations. This contextual information becomes crucial when reviewing results weeks or months later.
You could also add \(\mathrm{\LaTeX}\) based math equations onto the plots for more visually appealing and professional, I showed a way to use a customized version of \(\mathrm{\LaTeX}\) with around \(600\mathrm{\ MB}\) rather than installing the whole package, suppose some of you may rely on cloud based \(\mathrm{\LaTeX}\) environment including Overleaf.
Daniel Seshi’s article3 shows a tip by putting and commenting the commands for generating figures within the \(\mathrm{\LaTeX}\) source file’s respective figure block, so that future updates would be easier.
% Generate with:
% python [script].py --arg1 val1 --arg2 val2
% at commit [hashtag]
\begin{figure}
% LaTeX figure code here...
\end{figure}I prefer to put all the arguments within a yaml file but I think the above comments could provide very important contexts during critial time period when you wants to generate an updated version of figure. The key here would be to leave traces so that future you could be able to run codes quicker.
PDF Reports with Metadata
Rather than scattered image files, generate PDF reports with embedded metadata. Create figures with matplotlib, add text annotations within figures, save as PDFs with metadata (author, experiment ID, date), and organize in a consistent directory structure. This approach creates self-contained documents that maintain their context even when shared. Although matplotlib can generate figures within windows, I’ve found that PDFs with embedded metadata are significantly more practical, especially on macOS where Quick Look makes browsing through results effortless. Beyond the visible text within figures, embedding additional metadata provides contextual information that becomes invaluable months later.
On macOS, Quick Look makes previewing these reports fast and efficient, allowing you to browse results without opening full applications.
Evaluation and Result Management
Tracking results across multiple experiments remains challenging. I’ve personally struggled with organizing the training and evaluation process effectively. Initially, I tried using markdown tables to track results, which worked well with git version control but proved inflexible when comparing experiments across different parameter sets.
Consider using structured logs with JSON/YAML files that follow a consistent schema. Databases work well too—SQLite for local projects, PostgreSQL for team efforts. Spreadsheets like Excel or Google Sheets offer flexible analysis and visualization capabilities that make comparison particularly intuitive, though they don’t integrate as seamlessly with git. Automated reports that generate markdown or PDF summaries after experiments complete can bring everything together. I’m still experimenting with different approaches here, as this remains one of the most challenging aspects of ML research management.
While git works well for code, structured data storage proves better for results. You gain query capabilities for finding specific experiment outcomes, sorting and filtering options to identify patterns, statistical analysis tools to quantify improvements, and visualization options that highlight meaningful comparisons.
Practical Workflow Example
These tips combine into a practical workflow that streamlines machine learning research. Start by creating a project with separated code, data, and configuration files. Initialize a git repository to track changes from the beginning. Define your baseline parameters in YAML configuration files that can be easily modified and versioned.
During experimentation, generate random experiment IDs to keep filenames clean, queue experiments with your scheduler to maximize resource usage, and log comprehensive information about each run. Monitor progress using tqdm in your terminal and track metrics with Tensorboard for visual feedback.
For analysis, create custom visualizations that highlight the most important aspects of your results and generate PDF reports that capture the complete context. When comparing multiple approaches, organize results in a structured format that facilitates meaningful comparisons and create visualization dashboards that help identify trends across experiments.
Conclusion
Effective ML research requires more than just algorithm knowledge. By implementing these practical tips for code management, experimentation, visualization, and evaluation, you can create a more efficient and reproducible workflow.
I’ve presented a comprehensive pipeline for machine learning research and development based on my personal experience. While there’s no one-size-fits-all approach, these practices have significantly improved my productivity and result quality. The key takeaway is to establish systems early—for code organization, experiment tracking, visualization, and evaluation—that scale as your research progresses.
What strategies have you found helpful in your ML projects? Share in the comments below!
https://dustintran.com/blog/a-research-to-engineering-workflow#references
https://danieltakeshi.github.io/2017/07/15/how-i-organize-my-github-repositories
ISQED anchor
Leave a Reply